It is imperative to check the validity of any comparative model, first in computro, later by experiments. It would be foolish to trust blindly a model. The first step is of course to check the quality of the 3D structures used as a template for the comparative model. Reports on all PDB structures have been compiled in PDBreport (http://www.cmbi.kun.nl/gv/pdbreport/).
New structures and 3D models can be checked with the Biotech Validation Suite for Protein Structures. Three different software packages can be run on the server:
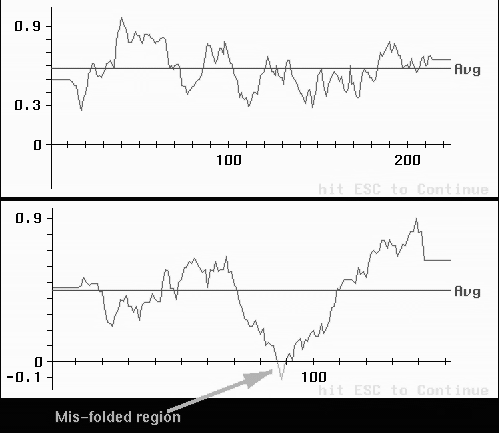
In addition, a 3D-1D profile verification can shed light on dubious areas in the structure. The 3D-1D server is at UCLA-DOE Structure Evaluation (http://www.doe-mbi.ucla.edu/Services/Verify_3D/).

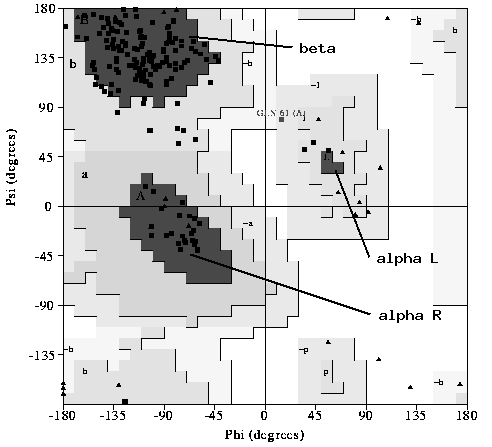
One of the most important checks for any 3D protein model is the Ramachandran plot. Any non-Gly residue outside of the 'allowed' zones should be carefully inspected.