5. Comparative Modeling:
From Sequence to 3D Model
5.1 Comparative Modeling: Principles
Comparative or homology modeling uses experimentally determined protein
structures as templates to predict the conformation of another protein
that exhibits amino acid sequence homology. Immediately, the question arises
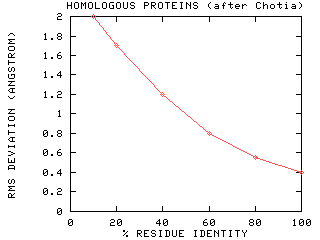
what the accuracy of such a method might be. A comparison of 32 homologous
proteins with known 3D structure gives an idea of the most optimistic estimate:
Flowchart for Comparative Modeling
- Find one or more proteins in the PDB that exhibit sequence similarity
to your protein
- Construct a sequence alignment
- Decide which regions are structurally conserved (SCR): helices, strands
- Construct loops either by analogy or from a database
- Place side-chains: use existing conformational info and rotamer databases
- Refine the model: packing, surface accessibilities, energy calculations
- Validate the model
Some hints
- Get the alignment right: no insertions/deletions in SCR
Example:
WRONG: papain .NQPV SVVLQAAGKD FQLY.
malprot .PLSV NV---GVNND FVAY.
actinidin .YQPV SVALDAAGDA FKQY.
**** ***** ** **** = SEC. STRUCTURE EL.
CORRECT: papain .NQPV SVVLQAAGKD FQLY.
malprot .VGPL SVNVG-VNND FVAY.
actinidin .YQPV SVALDAAGDA FKQY.
**** ***** ** **** = SEC. STRUCTURE EL.
Loops:
- some loops, for example beta-turns, have been very well characterized
- when borrowing loops include also information about the residues just
before and after the loop
- check the phi/spi angles of borrowed Gly residues
Usually, side chain torsion angles are conserved (observed by Karplus
& Sternberg)
Energy refinement can never fix a poor starting model. Think of energy
refinment as cosmetics
For more info:
- C.S. Ring & F.E. Cohen (1993). Modeling protein structures:
construction and their applications, FASEB J. XX, 783-790.
- M.C. Peitsch & J.T. Tschopp (1995). Comparative modeling of
Fas-ligand and other members of the TNF-family, Mol. Immunology 32,
761-772.
 5.2 Comparative Modeling: Servers and Freeware
5.2 Comparative Modeling: Servers and Freeware
© Copyright 1997 Christophe Verlinde
contact: verlinde@gouda.bmsc.washington.edu; tel: (206) 543 8865;
fax: (206) 685 7002


![]() 5.2 Comparative Modeling: Servers and Freeware
5.2 Comparative Modeling: Servers and Freeware