Molecular Machines
Section I 2.1 Type II Secretion System of Vibrio cholerae

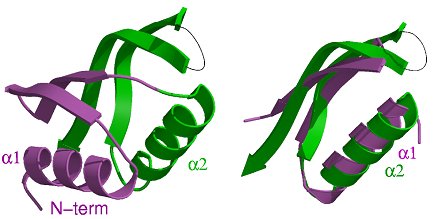
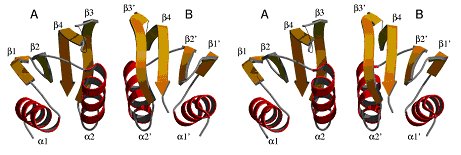
The structure of the periplasmic domain of EpsM from V.
cholerae. Upper: The structure of the dimer in stereo view. The most
likely physiological dimer is depicted with its non-crystallographic two fold
axis running vertically. The flexible loops are indicated by thin lines. EpsM
consists of a sandwich of a four-stranded antiparallel β-sheet and two
helices. Helices α2 and α2* mediate most of the intermolecular contacts,
the putative ligand binding groove is located between β3 and β3*.
Lower left: The monomer consists of two αββ-subdomains,
which have a very similar structure. Lower right: The superposition of
the two subdomains in the same orientation as of the green subdomain in (b).
(Jan Abendroth)
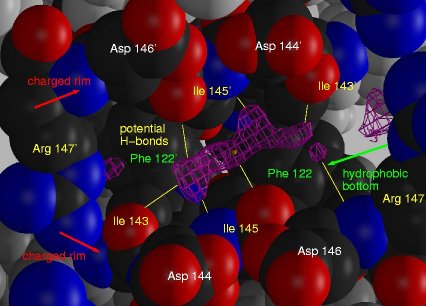
Extra electron density in the dimer interface of EpsM. A view into
the cleft between the two monomers along the 2-fold crystallographic axis
(marked with the yellow dot). The extra density 2mFo-DFc-density
at 1σ (magenta) is in hydrogen bond distance (yellow bonds) to Ile143,
Ile145, and Arg147 from both subunits and sandwiched between a hydrophobic
bottom and a charged rim. Residues involved in the cleft are shown in CPK-coloring,
all other in grey.
(Jan Abendroth)
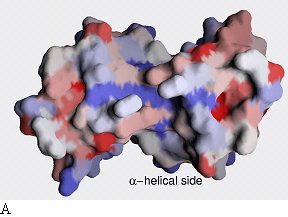
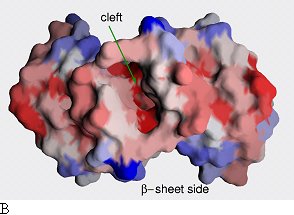
Sequence conservation plotted onto the surface of the
EpsM dimer. Two different sides are shown. Red color indicates high sequence
conservation, blue indicates low sequence conservation. Whereas there is little
sequence conservation on the surface of α-helical side (a), the surface
of the β-sheet side (b) and especially the cleft in between the β-sheets
have patches of high sequence conservation.
(Jan Abendroth)
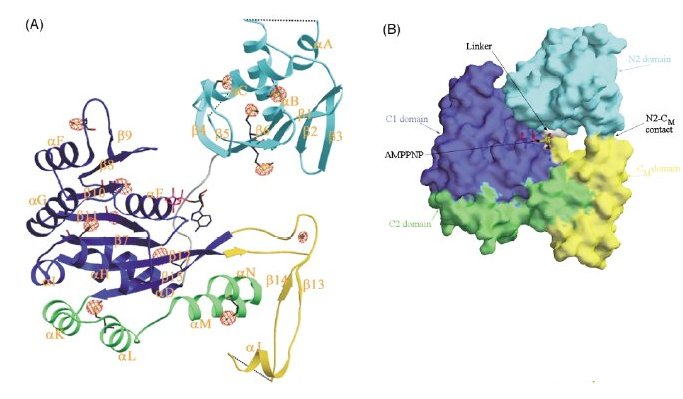
Structure of the EpsE monomer.
(A) EpsE ribbon structure with domains colored as follows: N2 cyan, C1 dark blue, CM yellow, C2 green. Due to loops with weak electron density, some residues between αA and αB, β4 and β5, and αJ and β15 (it looks like β14 in the diagram) could not be incorporated into the final structure. Hence, these loops are shown with dotted lines. The position of a bound molecule of AMPPNP observed in the 2.7 Å dataset is indicated. The position of eleven selenomethionine residues and the metal site identified by SOLVE are also shown, together with the anomalous Fourier map contoured at +4.
(B) Surface representation of the EpsE monomer and bound AMPPNP,
showing the positions of the domains and subdomains, with N2 colored cyan,
C1 dark blue, CM yellow and C2 green. The small ~300Å2
contact between the N2 domain (cyan) and CM subdomain (yellow)
is seen in the foreground. The surface of the linker residues (white) is visible
behind the nucleotide, AMPPNP.
(Mark Robien)
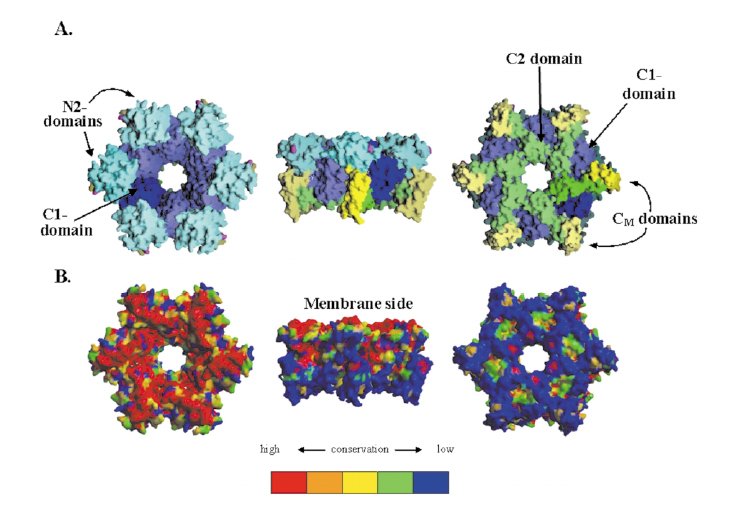
Hexameric ring model of EpsE.
(A) View from the proposed membrane-facing side(A1), side view(A2) and the cytoplasmic face(A3) of the hexameric ring model of EpsE constructed as described in the text. One monomer is shown with the domains colored as follows: N2, cyan; C1 dark blue; CM yellow; and C2 green. The other five monomers are colored in a lighter shade of the same colors.
(B) Same views as in (A1-A3), but with the surface colored
by sequence conservation.
(Mark Robien)
Section I 2.2 Trypanosomatid RNA Ligase from the Editosome
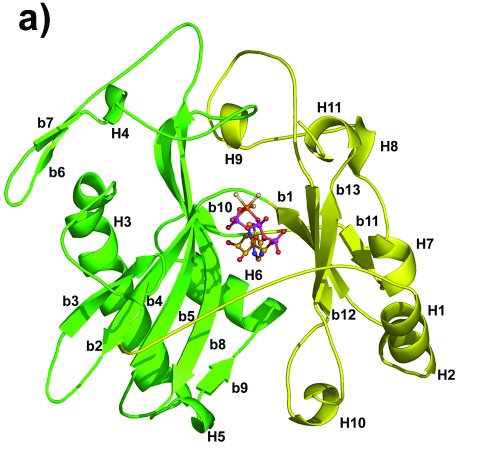
Overall structure of the adenylylation domain of TbREL1.
The two subdomains of TbREL1 are colored in green and yellow, respectively.
Helices are numbered as H1-H11 and β-strands are numbered as b1-b13.
(Junpeng Deng)
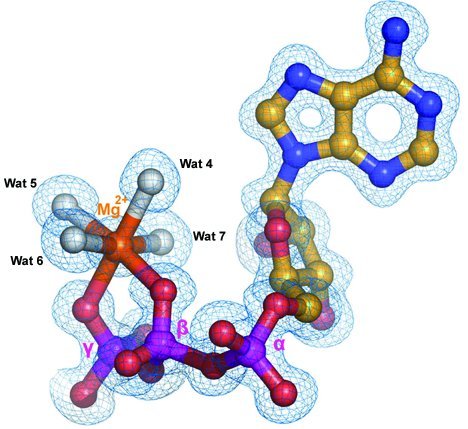
Interactions of Mg2+-ATP with TbREL1. The
2mFo-DFc electron density map covering
the ATP molecule and Mg2+ at 1.2 Å resolution. The Mg2+
is shown as an orange sphere. The direct water ligands of Mg2+
are shown as white spheres.
(Junpeng Deng)
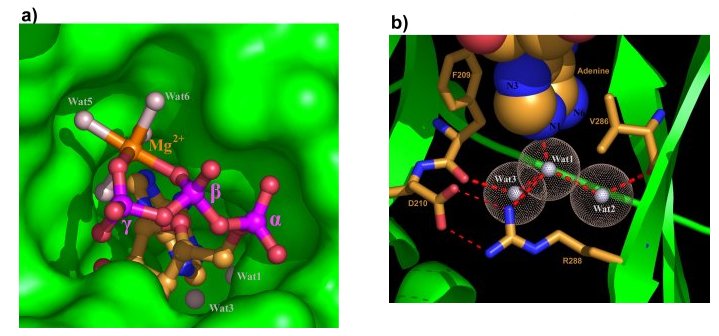
The ATP pocket.
(a) Mg2+ - ATP in its deep pocket with the protein surface in green.
(b) The three water molecules around the deep end of the adenine
pocket. Water molecules are shown as white spheres with dotted surface. The
adenine base is shown as space-filling model.
(Junpeng Deng)